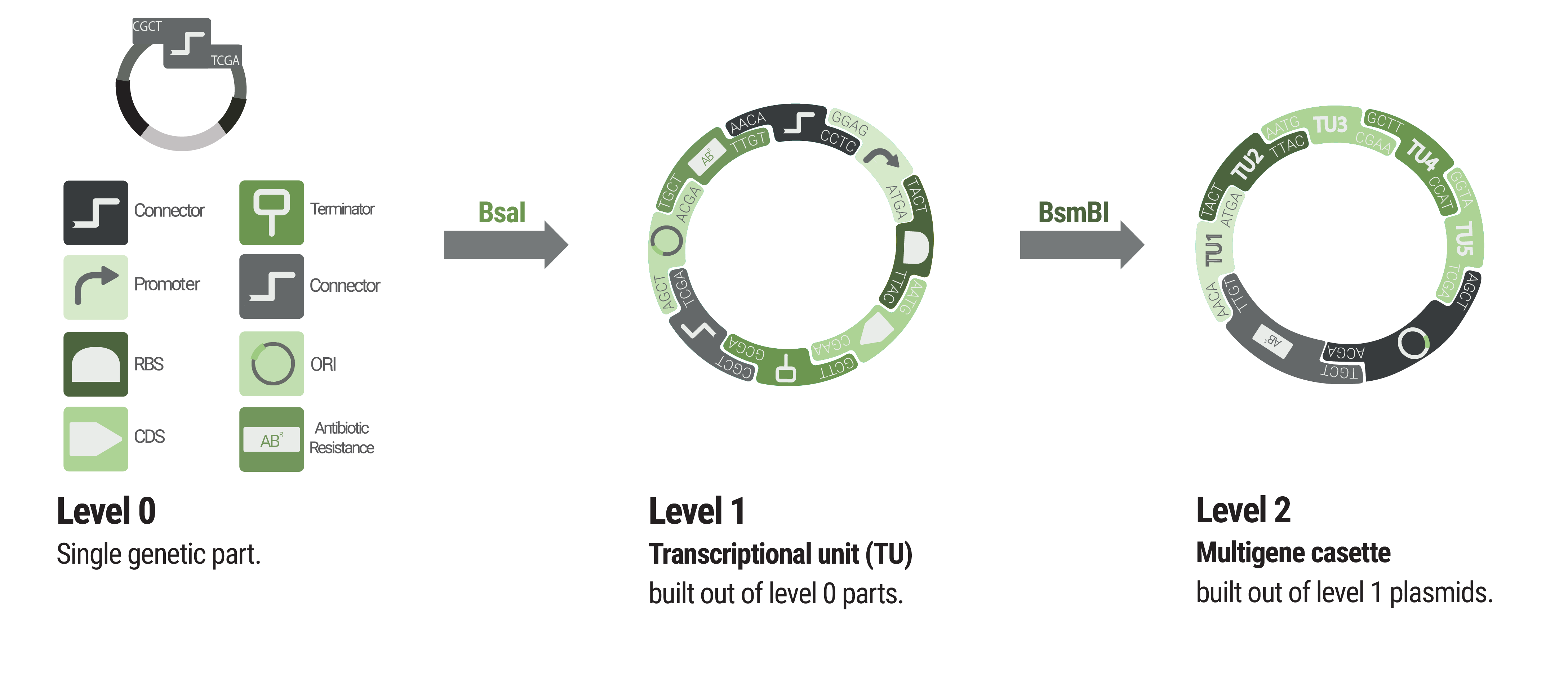
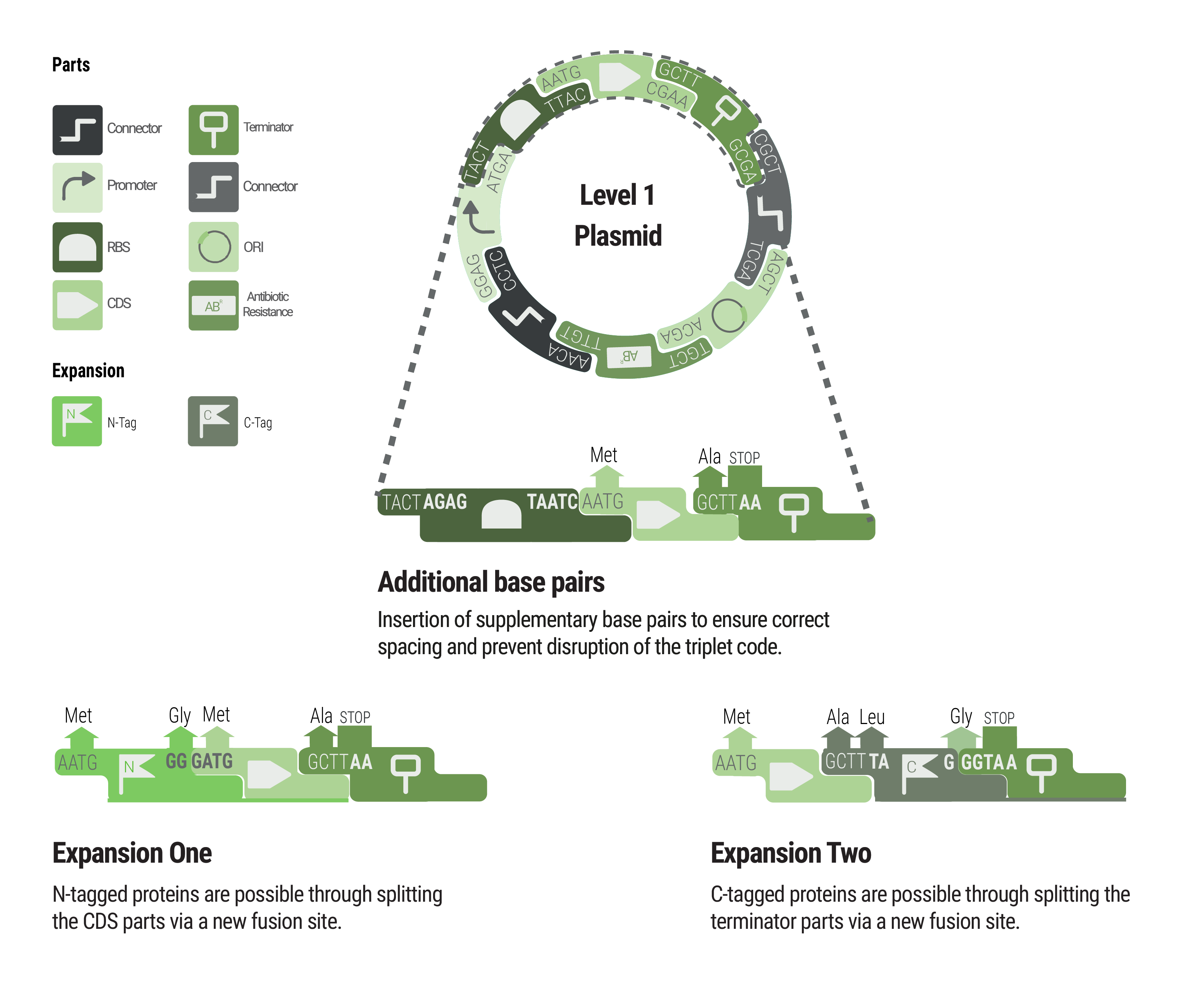
Overview
Origins of replication (Oris) are genetic elements
where DNA replication is initiated. In plasmids the
Ori sequence is responsible for it’s maintenance and
for the copy number inside the cell
(Selzer et al., 1983;
Brantl, 2014).
The origins of replication colE1, pMB1 and p15A
belong to the same family. They do not code for any
enzyme but are replicated by the hosts RNA polymerase
(Cesareni et al., 1991;
Brantl, 2014). The
polymerase transcribes a region 508 bp upstream the
Ori sequence (Tomizawa & Itoh, 1981;
Selzer et al., 1983)
synthesizing a pre-primer RNA called RNA II. During
transcription the RNA II underlies conformation changes
building secondary structures(Brantl, 2014). This
structures contain typical loops (Cesareni et al., 1991)
that binds to the plasmids’ Ori sequence building an
RNA-DNA hybrid (Cesareni et al., 1991; Brantl, 2014).
The RNA II is than cleaved by the hosts RNase H to
become a mature primer (Cesareni et al., 1991; Brantl, 2014).
Characterization
For our collection we characterized three Oris commonly
used in molecular biology: colE1, pMB1 and p15A. We
measured two different plasmids, one with and another
without a LUX cassette. Both plasmids consist of a
kanamycin resistance cassette and one of the three
Oris described. The LUX expression plasmid contained
a constitutively expressed LUX cassette of ~6kb. The
other one contained a connector sequence to build an
‘empty’ plasmid. By comparing this constructs you may
consider that the copy number is not only influenced
by the LUX expression but also by the plasmids sizes.
This Oris belong to the same family differing in
mutations in the RNA I region (Tomizawa & Itoh, 1981; Selzer et al., 1983).
We measured the plasmids’ copy number by qPCR using
the absolute quantification method.
A qPCR is set up the same way like a normal PCR but
with addition of a DNA binding fluorophore in this
case SYBR Green. SYBR Green binds double stranded DNA
emitting a high signal while unbound SYBR Green shows
only low fluorescence (Zipper et al., 2004).
In every PCR cycle the number of double stranded DNA
is duplicated emitting an increasing fluorescence
signal. This signal is detected after every cycle by
the qPCR machine and the value is saved. After the run
finished, normally after ~40 cycles, a signal threshold
is determined and the corresponding cycle when the
threshold was reached is saved for further analysis.
For the qPCR run first total DNA from our host containing
the plasmids of interest was isolated in the exponential
phase (OD600 ~ 0.5), purified using the
innuPREP Bacteria DNA Kit from Analytik Jena and all
samples normalized to ~5ng/ul with the Qubit fluorometer
from ThermoFisher scientific. Subsequently a dilution
series was made in 1.5ml tubes diluting the DNA 7
times 1:2. This way the dilution series contained 8
steps reaching from 20 to 2-7.
Two different primer pairs were used for the analysis:
one matching the housekeeping gene dxs present
once on the genome and the other matching the kanamycin
resistance cassette on the plasmid. The DNA samples
used for the amplification of the kanamycin cassette
were the same used for the dilutions 2-4
and 2-5. The threshold cycles (Ct) acquired
in triplicates from the dxs sequence were used
for a standard curve. By comparing the Ct values from
the resistance cassette with the corresponding standard
curve the number of copies could be determined as multiples
from the dxs sequence. It should be considered
that the dxs sequence is coded on the first
chromosome of V. natriegens at ~ one o’clock.
Due to that probably the sequence is present more than
once because of multifork replication of the genome.
To build the standard curve the Ct values were plotted
on the y-axis and the corresponding dilution steps on
the x-axis. The x-axis was set logarithmic and the
standard curve was calculated with Excel. The curve’s
formula was than used to calculate the corresponding
x-value from the resistance cassette’s Ct values.
Because the x-values describe a theoretical dilution
the Ct values were multiplied with this value and
with their corresponding dilution to obtain the final
amount of multiplies compared to the genome.
For every Ori an own standard curve was calculated.
Result
In our experiments we showed that the plasmids’ copy
number controlled by three different Oris differ a lot
when comparing V. natriegens with E. coli.
One possible explanation might be different expression
levels of RNA I and RNA II respecting the rate of
RNA I – RNA II bounds (Cesareni et al., 1991)
due to the divergent metabolism in V. natriegens
and E. coli. Another plausible explanation might
be the different methylation patterns in both organisms
probable affecting the formation of the RNA II secondary
structures and subsequently its binding affinity to
the DNA (Russell & Zinder, 1987; Cesareni et al., 1991).
It was shown that mutations especially in the loop I
structure might be responsible for Ori compatibility
and copy number control (Selzer et al., 1983;
Cesareni et al., 1991). The copy number is
mainly determined by two factors: the binding
efficiency of the RNA II to the DNA – specially
controlled by the stabilization of stem-loop IV –
(Cesareni et al., 1991) and the interference
of the complementary RNA I to the RNA II pre-primer
(Brantl, 2014). The RNA I is transcribed constitutively
from the complementary strand from RNA II pre-primer
(Brantl, 2014). Binding of RNA I to RNA II prevents
the correct folding of the pre-primer (Brantl, 2014).
This way the RNA-DNA hybrid can not be formed and
subsequently the primer maturation can not take place
(Brantl, 2014).
The columns show the average of the calculated multiplies for the different plasmids. The blue columns show the numbers for the plasmids containing a ~6kb LUX cassette. The orange columns show the numbers for the ‘empty’ plasmids without reporter. For every column six measurements have been calculated. Looking at the ‘empty’ plasmids it is clearly shown that colE1 and p15A remain high copy plasmids like in E. coli with a copy number of ~200 copies per cell. For pMB1 the copy number is scaled down becoming a low copy number Ori in V. natriegens. Looking at the LUX plasmids it is clearly shown that the colE1 Ori remains at a high copy number while pMB1 and p15A drop down to a significantly lower level.